Our platform
In this post, I would like to introduce the Humane Genomics Artificial Virus Platform.
Why develop an artificial virus platform?
For decades, a lot of research has been conducted to see if natural viruses can be used as cancer therapeutics, with limited success. Over the last 10 years, the technology to engineer, to build viruses specifically as cancer therapeutics has matured. However, most technology approaches to make these viruses are complicated, error prone, time consuming and expensive. To address these issues, we are using the latest technology, based on synthetic biology, to build our platform.
The Platform
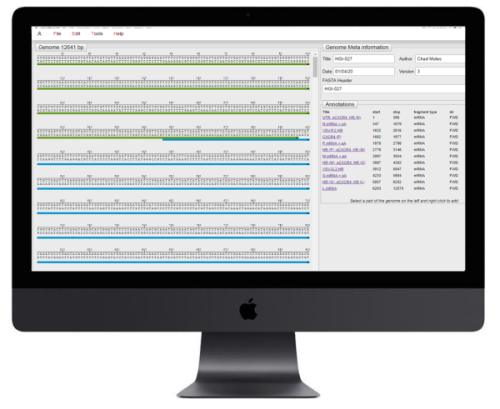
Our approach is to design every virus, or component of the virus, on the computer using our own proprietary virus DNA editor. Our editor enables us to easily add new elements, combine existing components and test if the design can be made using DNA synthesis.
After we complete and validate the design on the computer, the DNA is synthesized. With current technology, it is not practical to synthesize the complete genome of a virus in one piece, yet. Instead, we split the sequence into parts, which are synthesized as DNA fragments and then assembled together. After assembly, production and sequence validation of the correctly assembled DNA, the virus is “booted-up” in production cell lines. After successful rescue of the virus, we filter, quantify, and sequence verify once more. At this point, we are ready to test the virus.
What are the benefits of our platform?
Fast and high quality
Our platform allows for fast iterations. We are currently capable of producing sequence verified, testable amounts of virus in less than 3 weeks after we complete the design. We expect to go faster still, with timelines under 2 weeks in the near future. In addition to the speed, our process also produces high quality results. We currently see 73% success rate from design to validated DNA.
Other approaches like restriction enzyme digestion, PCR amplification and CRISPR take considerably more time (up to 6 weeks per design change) and have a much higher chance of failure. This means several attempts are needed (on average 7) to make one successful edit, further increasing cost and timelines.
Unprecedented capabilities
Using our platform, we can engineer viruses, which are safe and effective. To do this, we use multiple markers to identify the cancer cells. These are surface markers (receptors) and internally present markers (proteins). Our viruses are designed to match a specific targeted receptor to infect only the cells of interest. Viral replication and killing of the cell is controlled by a switch, which only allows replication in the presence of a specific protein. We can also use a switch to regulate the production of a specific protein to further enhance the therapeutic effect, but again, only do this in targeted cells.
The decreasing cost of synthetic DNA combined with the speed and efficiency of our platform means we can try very complicated designs several times and still make them work in relatively short periods of time and on a small budget. To name a few examples of the designs we have successfully tested: multiple glycoproteins on a single virus, gene rearrangement and genetic circuits. Next to complicated design concepts, we use our own software to codon optimize existing genomes without changing functionality. Codon optimization can drastically improve the replication speed of a virus making it more effective as a cancer therapy.
Taken together, our approach leads to unprecedented therapeutic potential.
Building a digital library
Since our platform is digital first, every virus (and components) we test, also exists digitally in our database. If we need a component or virus again, we can synthesize an exact replica on demand. Combined with high standardization of our lab processes, this leads to high reproducibility of results. Through our work, we are establishing a library of verified virus designs and reusable virus design components. Building on this, it becomes increasingly easier to engineer the next virus for a new type of cancer.
Today, our library already contains 9 different glycoproteins, which are tested and confirmed to target some of the most well understood cancer specific receptors like HER2, EGFR, GPC3 and others. Our library also contains 4 verified replication switches and therapeutic genes. All of these components have been tested and validated, and can be reused in the creation of new cancer killing viruses.
Using our platform we are now working on our first oncolytic virus to treat Hepatoblastoma. We hope and expect to add many more in the near future.