Regulating viral replication using RNA aptazymes
The biggest problem in cancer therapeutics is selectivity. Using oncolytic viral therapies, high selectivity can be achieved using the combination of selective infection and selective replication. Here, we want to address selective replication. For DNA viruses, this can be accomplished with promoters to control transcription and genome replication. However, for RNA viruses, this does not work. A potential solution to control replication for RNA viruses is the use of aptazymes.
Aptazymes are self-cleaving units composed of a riboswitch and an aptamer. Aptamers are single-stranded sequences or oligonucleotides that bind a specific target protein. In case the target of the aptamer is present, the riboswitch will either turn “ON” or “OFF” depending on the design.
Aptamers have several favorable properties. First and foremost, they exhibit greater binding affinity than antibodies. Protein detection of zeptomole amounts have been reported with aptamers, that’s 10^(-21) mol. Also, they are significantly smaller, often 20-60 base pairs in length. For comparison, Trastuzumab (FDA approved HER2 antibody) is 1,992 base pairs long. Taken together, this makes them great to use in aptazymes to regulate functionality.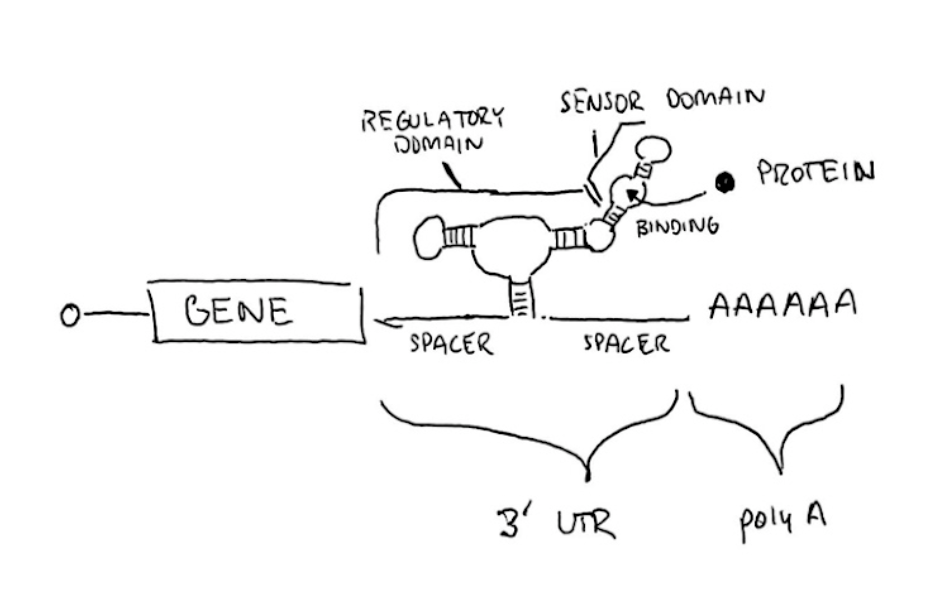
The drawing shows an “ON” switch design. A riboswitch that will automatically cleave is used. When it cleaves, the poly-A sequence is removed from the gene. As such, the RNA sequence becomes unstable and is degraded by the host cell. When the target protein binds the aptamer, there is a structural change in the riboswitch, which inhibits the cleavage activity. The poly-A sequence remains coupled to the gene, ensuring stability and translation of the RNA.
Most aptazymes that have been developed thus far use antibiotics or drugs, such as tetracycline or theophylline, as the “protein” to control the ON activity of the gene. These basic science studies have laid the groundwork for this technology and approach.
At Humane Genomics, we are focused on moving these scientific advances from the bench to the bedside. We’re taking advantage of the modularity of the sensor domain, and are using aptamers that target cancer-specific proteins.
Up to 90% of cancers express telomerase reverse transcriptase (TERT). This makes TERT a great target for an aptazyme “ON” switch. We coupled the ON switch to a gene coding for a Green Fluorescent Protein (GFP). We then added the construct to cells that express TERT (bone cancer cell line called U2-OS) and measured fluorescence, which is an indication of ON activity or gene expression. To evaluate the specificity of our aptamer, we added a TERT antibody to some cells to bind the protein and prevent it from binding our ON switch. The results are shared below.
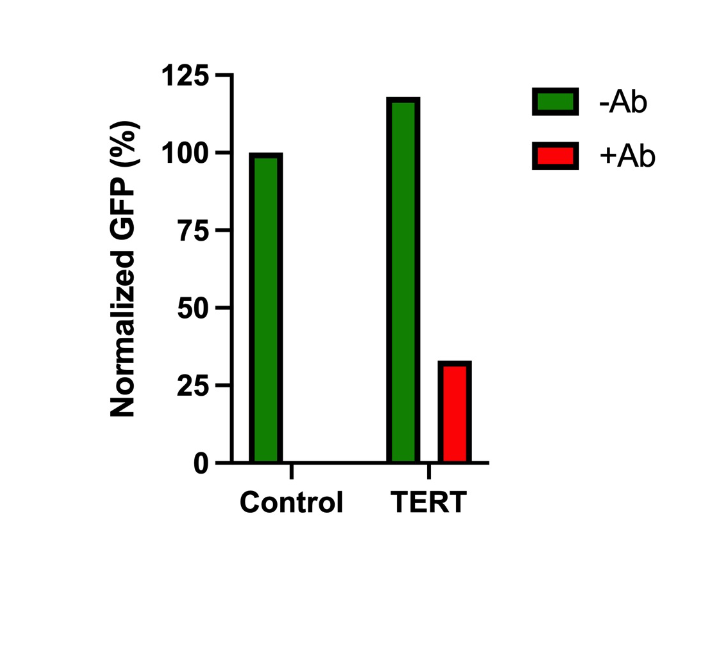
We plan to incorporate the TERT ON-switch, discussed here in some of our oncolytic viruses. Separately, we have developed aptazymes for other cancer specific protein targets.
Takahashi et al. engineered an aptazyme that recognizes guanine as an OFF switch (binding of the protein causes a structural change that enables cleavage activity) and had success with reducing viral replication of VSV (image below).
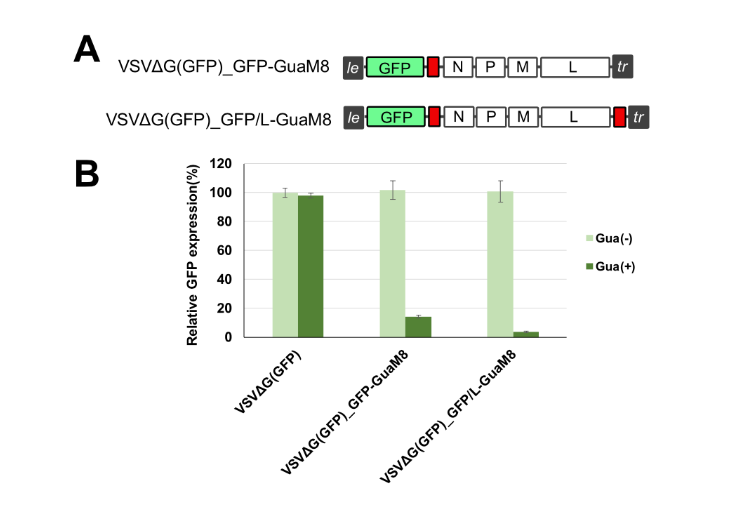
We will explore this concept to create a safety switch for our oncolytic viruses. We look forward to sharing these results with you soon!